Overview and Features of SBTOOLBOX2
| The Systems Biology Toolbox 2 (SBTOOLBOX2) for MATLAB offers systems biologists a powerful, open, and user extensible environment, in which to build models of biological systems. Experiments can be performed on models, just like in real lab life but in silico. The representation of models, experiments, and measurement data is intuitive and easy to use. The toolbox features a wide variety of specialized analysis tools and MATLAB adds to that by a large number of inbuilt functions and a high level scripting language, allowing the user to quickly and efficiently add new functionality. |
Main Features SBTOOLBOX2
|
Model Representation
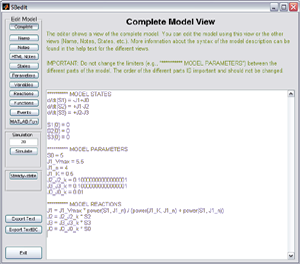
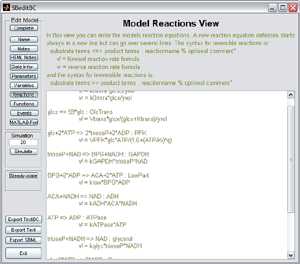 Define your models using a simple syntax based on differential equations or based on biochemical reaction equations. To see examples
you can click on the figures to the right.
Define your models using a simple syntax based on differential equations or based on biochemical reaction equations. To see examples
you can click on the figures to the right.Additionally it is possible to define differential equations, initial conditions, variables, and reactions using a vectorized syntax. An example for which this syntax is useful is:
d/dt x[n] = kon*x[1]*x[n-1] + koff*x[n+1] - (kon*x[1]+koff)*x[n]
SBML Enabled
The toolbox supports the import and export of SBML models. On this page you can find information about its SBML compatibility.
Experiment Representation
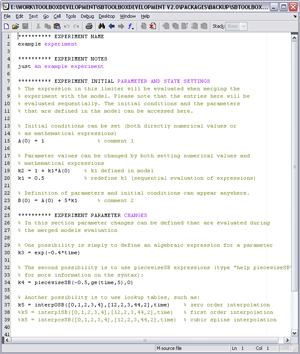 In order to match the in silico implementation to the real workflow in a lab the toolbox allows to describe in silico experiments
that are to be performed on the model. Click on the figure to the left to see an example experiment description.
In order to match the in silico implementation to the real workflow in a lab the toolbox allows to describe in silico experiments
that are to be performed on the model. Click on the figure to the left to see an example experiment description.
Experiments and models can then be merged to obtain a simulatable model that reflects the defined experiment. Especially in the case where several
different experiments are performed on the same system it is very useful to be able to only keep one copy of the model and to code the different
experiments per se.
Measurement Representation
Measurement data collected during experiments can be imported to the toolbox, visualized, manipulated. Both Excel and comma separated value (CSV) formats are accepted.
Simulation, Analysis, etc.
 The toolbox features simulation (both deterministic and stochastic) and a wide range of analysis tools, such as:
The toolbox features simulation (both deterministic and stochastic) and a wide range of analysis tools, such as:
- Steady-state and stability analysis
- Stoichiometry
- Parameter sensitivity analysis
- Determination of moiety conservations
- Localization of mechanisms leading to complex behaviors
- Statistics (clustering, percentiles, boxplots, etc.)
- Global and local optimization algorithms
- etc.
Additional Features
Additionally, the toolbox contains many auxiliary functions that are useful for facilitating the access to model or data elements, writing own scripts, and last but not least to make it independent of other commercial MATLAB toolboxes and third party packages.
Documentation
| Section | Description |
|---|---|
| User's Reference | Complete user's reference guide. All functions of the toolbox are explained in detail and by examples. |
| SBML Compatibility | Important notes about the compatibility of the toolbox with SBML models. |