Overview and Features of SBPOP
| The SBPOP extension package (SBPOP) for the Systems Biology Toolbox 2 adds functionality related to pharmacometrics and systems pharmacology applications. Functions are available that support the complete model building workflow for population PK models. Additionally, SBPOP contains interfaces to Monolix and NONMEM, powerful and flexible graphical analysis functions, and a user-friendly and powerful support for clinical trial simulations. |
Main Features SBPOP
|
Dosing descriptions and model syntax
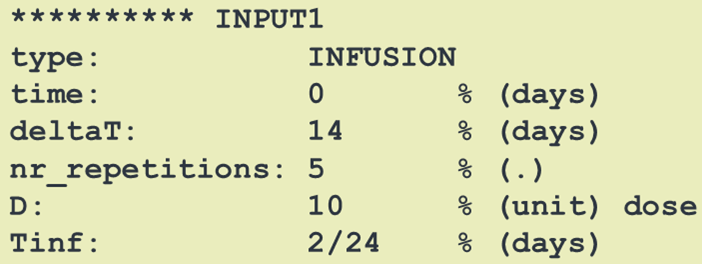 Dosing descriptions (SBPOPdosing objects) are used to define the type of administration of doses to a model. Possible dosing administrations are infusion, bolus, first and zero order absorption.
Each dose administration needs to be given a name "INPUT*" where "*" is a numeric value. This name then can be used within an SBmodel to apply a certain dose input to a desired
differential equation / compartment.
Dosing descriptions (SBPOPdosing objects) are used to define the type of administration of doses to a model. Possible dosing administrations are infusion, bolus, first and zero order absorption.
Each dose administration needs to be given a name "INPUT*" where "*" is a numeric value. This name then can be used within an SBmodel to apply a certain dose input to a desired
differential equation / compartment.
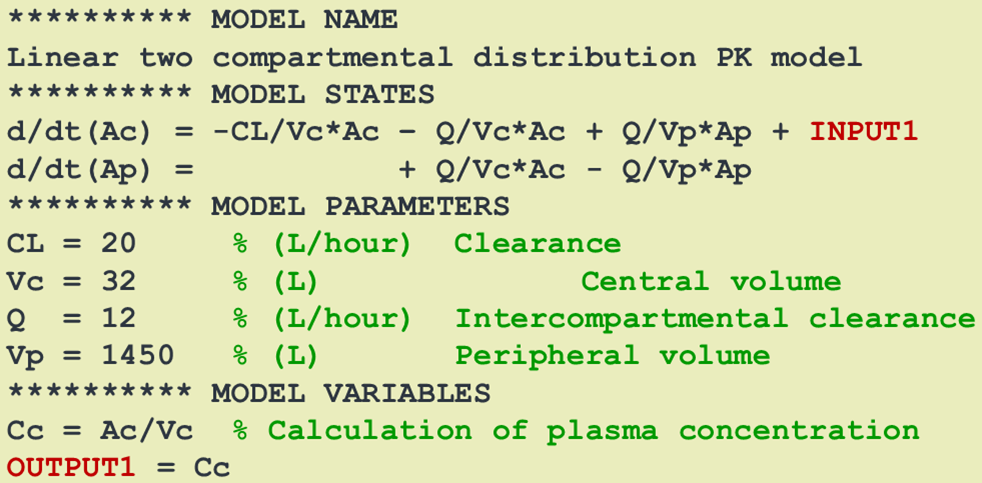 In an SBmodel, the INPUT* identifier is used to select the differential equation (or compartment) to which the dose should be added. The syntax is flexibel and powerful. Several
dosings/inputs can be defined and applied to different compartments. Doses can also be split up between compartments.
In an SBmodel, the INPUT* identifier is used to select the differential equation (or compartment) to which the dose should be added. The syntax is flexibel and powerful. Several
dosings/inputs can be defined and applied to different compartments. Doses can also be split up between compartments.
The "OUTPUT*" definition of variables is used for identification of the model outputs that are to be considered for parameter estimation with the NLME tool of choice.
Automatic generation of NONMEM control files, MLXTRAN models and Monolix projects
The SBmodel syntax can automatically be converted to MLXTRAN models and NONMEM control files. This of course is limited to ODE based models, which is the focus of the whole SBPOP PACKAGE. The only things that need to be defined apriori are an SBmodel and an SBPOPdosing object.Additionally, SBPOP allows to generate complete Monolix and NONMEM parameter estimation projects. The typically used settings can be manipulated directly from within SBPOP, without the need to start the Monolix GUI or to write Monolix project or NONMEM code manually. Obviously, this allows for powerful scripting of many different NLME estimation based analyses.
Graphical analysis functions
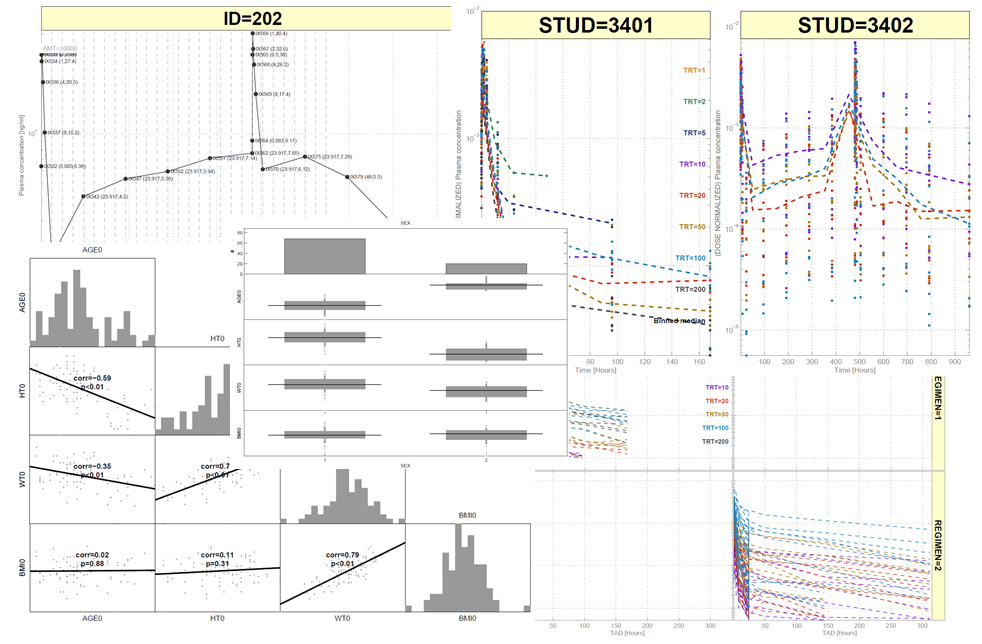 SBPOP contains many powerful functions for graphical data analysis. Unfortunately Mathworks has so far not seen the value in Trellis and Facet-Grid plots (other than in the sysbio toolbox),
so these are now implemented in SBPOP and somehow tuned to look similar to R plots. Additionally, support for grouped histograms, etc., is available.
SBPOP contains many powerful functions for graphical data analysis. Unfortunately Mathworks has so far not seen the value in Trellis and Facet-Grid plots (other than in the sysbio toolbox),
so these are now implemented in SBPOP and somehow tuned to look similar to R plots. Additionally, support for grouped histograms, etc., is available.
Population PK workflow
SBPOP contains extensive functionality targeting population PKPD analyses. At the backend Monolix and/or NONMEM are used to perform NLME parameter estimation.Population PK analyses often are very time consuming. Standards enable these analyses to be performed with considerably increased efficiency. SBPOP assumes a certain data standard, covering not only for population PK, but also PKPD and time-to-event analyses. No need to request several different datasets from the programmers.
Functionality is included that supports a complete population PK model building workflow. The "Pareto principle" has been applied, meaning that this workflow is not required to be able to cover 100% of the possible population PK models, but if it covers 80% and does so efficiently, i.e., a lot of time is gained and can be put to better use.
Clinical trial simulations
The core of the clinical trial simulation functionality is the parameter sampling function. It samples population parameters from the uncertainty distributions and individual parameters from the variability distributions, directly from the Monolix and/or NONMEM results - easy to use and powerful. The user has full control over what exactly to sample. Different types of distributions of individual parameters (normal, log-normal, logit-normal, etc.) are taken into account and handled automatically, estimated correlations of fixed and random effects are also handled automatically. The sampling function is also able to handle user defined covariate settings automatically.Once the parameters for a simulation can be easily sampled, the clinical trial simulation boils down to repeated simulations, which can be easily and efficiently implemented using the SBPOP package.
Documentation
| Section | Description |
|---|---|
| User's Reference | SBPOP user's reference guide. |